BNT162b2v8/v9 - a case of plausible deniability
high stakes carny tricks and the people who fall for them
For all the reading I do, you’d think I’d be better at it. Pustekuchen! Beyond the primary finding of ~3-5x more functional neutralizing antibodies1 induced by V9 compared to V8, the TGA Nonclinical Evaluation Report also tells us this:
V8 and V9 have identical amino acid sequences of the encoded antigens and differ only in their codon optimisation sequences, which were designed to improve antigen expression (V9) with higher content of cytosine ribonucleotides.There is no evidence of the RBD variant BNT162b1 having “V8” and “V9” versions. It encodes antigen version 5, hence the name V5modRNA. Does the shorter length of the RBD mRNA (1262 nucleotides) vs the 2P-spike (4283 nt) play a role?
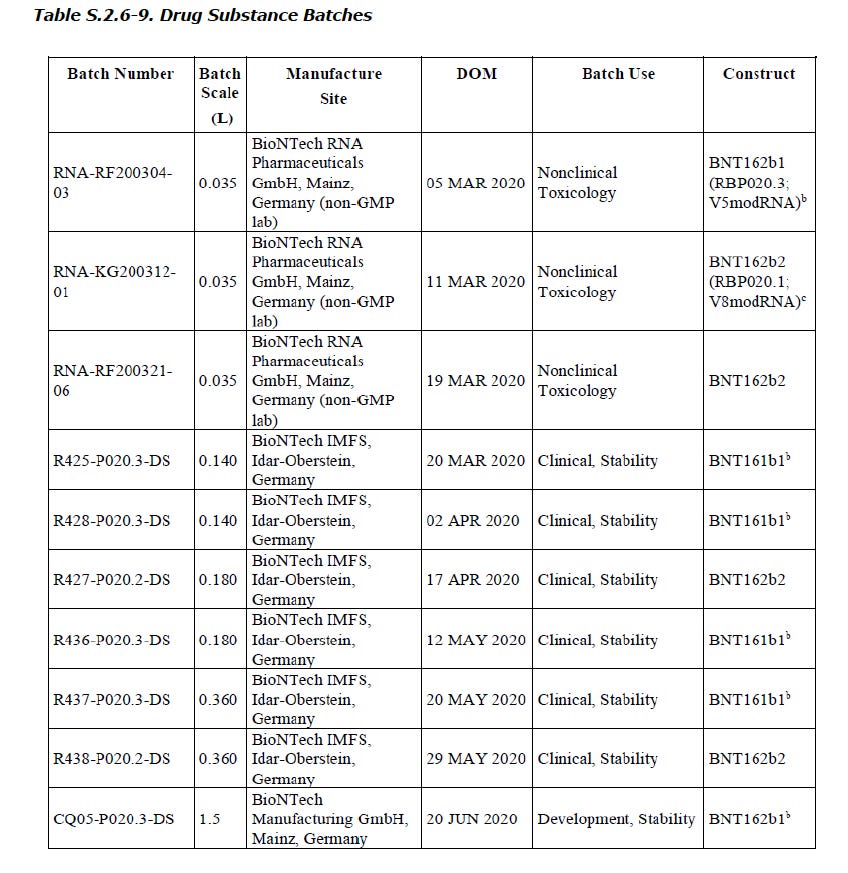
This document (thanks @canningpharm) from the covidtruths EMA leak gives a lot of pertinent information. Firstly, my prediction for production date of BNT162b2v8 was correct: “This still leaves the margin incredibly, if not impossibly, close: b2v8 DS release can be assumed to have occurred somewhere between March 5-9 and begin of b2v9 DS production was (March 19 minus ~1 week) somewhere around March 12.”
The BNT162b2v8 date of manufacture was March 11. The very next batch of mRNA to go into production was BNT162b2v9, drug substance released March 19. This means there was no research that could have occurred in between, and have informed the decision to change the codon-optimization. This in turn means that the intention was always to use the v9 antigen. This poses the question, why even make v8?
I suspect the answer lies in study 38166. I’ve already written a little about the batches from the future time-and-date paradoxes concerning the used material. It seems to be a little more sinister though. The BNT162b2v9 produced March 19 was finished product from Polymun on April 9, 15 days after the other non-clinical material (a1, b1, b2v8, c1).
In other words, they only made BNT162b2v8 for the repeat-dose toxicity study 38166, because they could have used v9 had they waited briefly. It is also noteworthy that BNT162b2v8 was made on March 11 and was finished product by March 26 (15 days), despite being processed together with BNT162a1, b1, and c1, while BNT162b2v9, made on March 19 with the same batch size as BNT162b2v8, took 22 days to be returned as finished mRNA-LNP product from Polymun. Was Polymun told to sit on the batch for an extra seven days in order to make the use of BNT162b2v8 in study 38166 less suspicious?
Both Moderna and Pfizer-BioNTech vaccine regulatory filings rely heavily on studies with “equivalent” products, inherently implying comparability of the platforms, which might explain why EMA/FDA didn’t bat an eye at the “last-minute” substitution of candidate for human dosing; “same” platform, “same” antigen, “just” ~3-5x more antibodies, which is good.
There’s really no excuse for BNT162b2v9 not having it’s own repeat-dose toxicity study with histopathologic examination before human dosing, and the errors in 38166 batch release/animal dosing dates/animal autopsy dates reinforce this notion, as they are too early. The few days between claimed animal dosings and BNT162b2v9 production however were entirely sufficient for regulatory oversight to give it the nod, instead of insisting on a re-do with the “improved” formulation, which would only have been a couple of weeks. Considering the first human doses of BNT162b2v9 were given on June 8, there was all the time in the world.
Instead, study 20GR142, the repeat-dose toxicity study involving BNT162b2v9 and BNT162b3c, only began on June 23. You can start reading the pdf from p549/603 onwards, as that’s where the important stuff is: the VisMederi virus neutralization report, the clinical pathology report (begins p561 and contains the hilarious sentence “Bone marrow smears were prepared for all animals. Bone marrow smear slides were stained with May-Grunwald Giemsa and were not examined.” ) and the anatomic pathology report (p578). The final reports were only done in December 2020, just before the EUAs started rolling in.
What is it about v9 they didn’t want to have on paper? Were they cautious of leaving the histopathology to an external lab? The only available histopathology on BNT162b2v9 is Pfizer-authored and reviewed. Why even make the effort? It’s absolutely unplausible they happened upon some stroke of genius while making v8 which led them to create v9, but then still used v8 in the toxicity study required to begin dosing humans, to no discernable reaction by regulators.
A recent Pfizer-BioNTech paper on studies 38166 and 20GR142, “Toxicological Assessments of a Pandemic COVID-19 Vaccine—Demonstrating the Suitability of a Platform Approach for mRNA Vaccines.” Vaccines 2023, 11, 417. describes v9 as “a codon-optimized version of BNT162b2 (V8), which encodes for
identical amino acid sequences but with improved protein expression and immunogenicity.”
Some more quotes:
”Note that there were two variants of the BNT162b2 vaccine tested. The RNA component of the two sequence variants (V8 and V9) have different nucleotide sequences, but both encode the same antigen.”
“Two versions of BNT162b2, variants 8 (V8) and 9 (V9), were evaluated non-clinically. These two variants differed only in their codon optimization sequences, which were designed to improve antigen expression; the amino acid sequences of the encoded antigens were identical.”
If they were so identical, why did they go out of their way to only report results that are tricky to be compared? pVNT90 and S1-IgG are reported for study 38166 with v8, pVNT50 for study 20GR142.
Then there’s also the curious issue of the test facilities. 38166 was conducted by “Laboratory of Pharmacology and Toxicology GmbH & Co. KG, Hamburg” and the immunogenicity assessment (including virus neutralization) was done by BioNTech. 20GR142 was conducted by Pfizer Groton Connecticut, yet the virus neutralization was done by VisMederi in Italy, who also did the same work for study 20256434, the fertility study by Charles River France. BioNTech used a VSV/SARS-CoV-2 pseudovirus, VisMederi used a real SARS-CoV-2 strain. BioNTech’s pVNT90 had an ULOQ of 2048, VisMederi’s was more than twice as high.
Here comes the kicker: for the R-20-0253 virus neutralization and antibody titre study that enrolled 60 b1 and 72 b2 patients from BNT162-01 in Germany, the VNT work was done by UTMB Galveston, and the antibodies by Pfizer Pearl River. Why not VisMederi on this side of the Atlantic? Why not in-house at BioNTech for that matter? (This study is the first of five in the bnt162 interim reports file mentioned in the last paragraph).
I’ll need to spend a lot more time reading and comparing the three studies 38166, 20256434 and 20GR142 in order to better pin down the differences between vaccine candidates. Study 38166 reads a lot more “honestly” than the Pfizer-authored 20GR142; fewer summaries and less spreading out of information into appendices and extraneous files.
There are still two unreleased studies concerning v8; perhaps my FOIA request to PEI will get them before they’re released to PHMPT. Fingers crossed, eh? Now I’ll try and get my write-up of the “bnt162 01 interim3 reports” PDF done before we get the next batch.
"BNT162b2 (V8) induced lower titre of functional neutralising antibodies (~ 3-5 fold) compared to BNT162b2 (V9).” P24/58 foi-2389-06.pdf







must be exhausting for them trying to keep everything hidden, it's way beyond my understanding so thank you for breaking it down
Well this is gold. Just reading your stack.
I have an idea why they had the toxicology for V8 and not V9. It is possible that V9 is making the spike protein and another peptide. The Western blot issues are trying to hide this.
And yes, the toxicology studies using V9 were a little concerning especially liver vacuolations, but data came in too late for them to stop the trials...my complaint with the rolling review. I dont think many people realize this.
Also in study 3866 they used a 100ug dose of V8!! Why do this study at all?
I think they are hiding what exactly that 162b2V9 is actually coding for. Its not the spike, or more accurately, not just the spike.